Studying splicing variants reveals the effects of genetic differences in pre-mRNA splicing between modern and archaic humans, and provides insights into how natural selection acted on splicing in human evolution.
This web page was produced as an assignment for an undergraduate course at Davidson College. Read more here.
Studying variation in hominin evolution can be critical for understanding genetic differences between modern and archaic humans. In a 2023 study, Rong et al. focused on splicing variants unique to Neanderthals and Denisovans (two groups of archaic humans) that are potentially pathogenic, as well as introgressed splicing variants that could account for variation in modern humans (Rong et al., 2023). Introgression is the transfer of genetic information between species due to hybridization between them, and introgressed variation in modern human populations can be used to understand the functional consequences of genetic variation between groups. Splicing is the process of introns being removed from pre-mRNA after transcription takes place, and therefore affects translation and gene expression.
In the past, modern humans coexisted with other hominins, including Neanderthals and Denisovans. Studying ancient genomes from these groups enables us to compare the genomes of modern and archaic humans, and has indicated that several admixture events occurred between modern and archaic humans. An admixture event is interbreeding between individuals from populations that were previously isolated. Archaic humans had smaller effective population sizes (the number of individuals that contribute to producing the next generation) than modern humans do, suggesting that more of their deleterious alleles were subject to purifying selection after introgression in the modern population. Purifying selection reduces genetic variation at sites that are under direct selection and at associated neutral sites, and plays an important role in shaping genomic diversity in populations (Harris & Nielson, 2016). When effective population size is small and genetic drift is strong, alleles that are weakly deleterious tend to remain in the population similarly to neutral alleles. Neanderthal exome sequencing has confirmed this, which provides direct evidence that purifying selection was weaker in Neanderthals than in humans (Harris & Nielson, 2016). Similarly, Juric et al. noted that most of the purifying selection against Neanderthal alleles can be attributed to acting on weakly deleterious alleles, and proposed that most of these alleles resembled neutral alleles in Neanderthals but later became selected against after entering modern populations (Juric et al., 2016).
Rong et al. found that in modern human populations, purifying selection acted against splicing variants while positive selection in adaptive introgression favored splicing variants. Adaptive introgression is the introduction of a new variant that increases the evolutionary fitness of the population receiving the variant. Compared to Neanderthals, modern humans had greater purifying selection on variants that disrupt splicing, primarily due to their larger effective population size. When considering splicing variants that have a moderate effect, there was positive selection for alternative splicing after introgression. This is likely due to increased purifying selection against introgressed variants that reduced the inclusion of exons, along with positive selection for moderate splicing variants. They also identified splicing variants contributing to variation in a range of modern human phenotypes, such as balding, hemoglobin levels, and lung capacity. Their findings provide insights into how natural selection acts on splicing in human evolution, and demonstrate how potential variants associated with differences in gene regulation and phenotypic expression can be identified.
Their experimental approach involved identifying 962 splicing variants that differed between modern and archaic humans using a massively parallel experiment. This type of experiment can be used to find causal variants that affect transcription, and they used a system called MaPSy to identify splicing mutations that influence exon recognition during pre-mRNA splicing. This type of system has been used to characterize the splicing impacts of several types of variants, and could be applied to genetic variation within modern and archaic humans.
They compared the variants obtained from MaPSy to computational predictions of different splicing effects that were generated by an AI system. They specifically examined fixed negative exonic splicing mutations that were associated with modern, archaic, and Neanderthal lineages. They also identified potential phenotypic impacts of splicing variants by exploring whether they were associated with trait variation. Finally, they identified seven variants across eight phenotypes that showed evidence of impacting both splicing and phenotype. However, six of these variants were missense variants, suggesting that other mechanisms were influencing phenotype since missense variants change which amino acid is encoded at that position.
Overall, this was a strong study because it provides insights into how natural selection plays a role in human evolution by acting on splicing variation. Using these approaches allows for opportunities to characterize the function of variants found in populations that may be extinct or underrepresented in the current literature. Improvements in current techniques for the functional interpretation of variants will also allow us to improve our understanding of human origins and phenotypes. However, limitations to this work are associated with the design of the experimental system. The authors noted low statistical power of the variants studied, which could have limited the extent of the evaluation of their results.
One question that remains is how these patterns may differ for intronic variants. There are intronic variants that can disrupt splice signals, which could be tested similarly to the approaches taken here. By extending this work to intronic variants, more complete data could be gathered about how selection acting on splicing variants contributed to human evolution. This work has profound significance because studying ancient human genomes and drawing comparisons to modern human populations can provide us with more information about the specific mechanisms of human evolution, as well as human migration patterns and links to anthropology. Additionally, this work could have implications for evolutionary medicine, which involves applying insights from evolution and ecology to biomedicine.
Works Cited
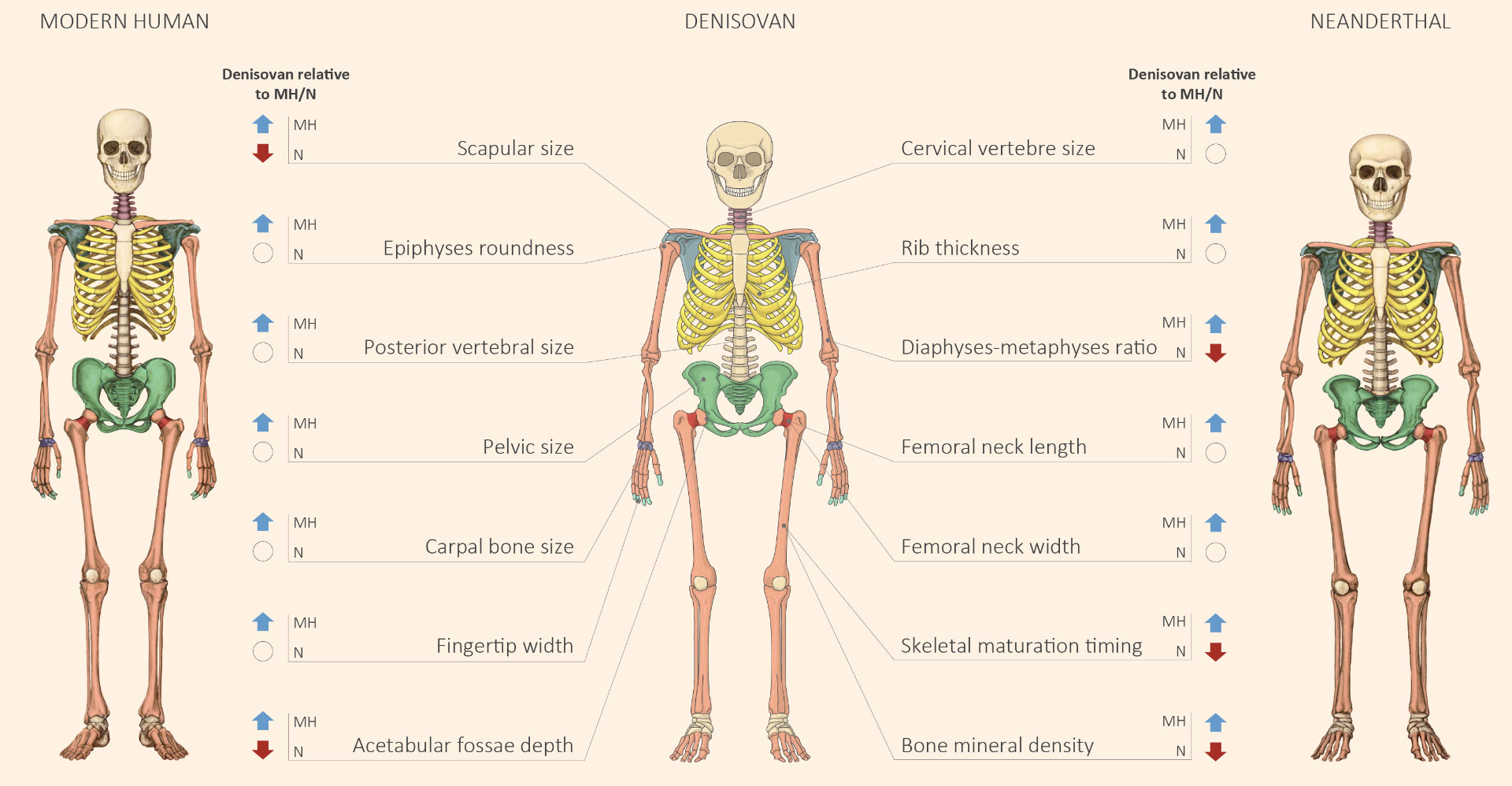
Harel, M. Anatomical comparison of modern human, Neanderthal and Denisovan skeletons. SciNews. https://www.sci.news/genetics/denisovan-anatomy-07610.html
Harris, K. & Nielsen, R. The Genetic Cost of Neanderthal Introgression. Genetics 203, 881–891 (2016). https://pubmed.ncbi.nlm.nih.gov/27038113/
Juric, I., Aeschbacher, S. & Coop, G. The Strength of Selection Against Neanderthal Introgression. PLOS Genetics 12, (2016). https://pubmed.ncbi.nlm.nih.gov/27824859/
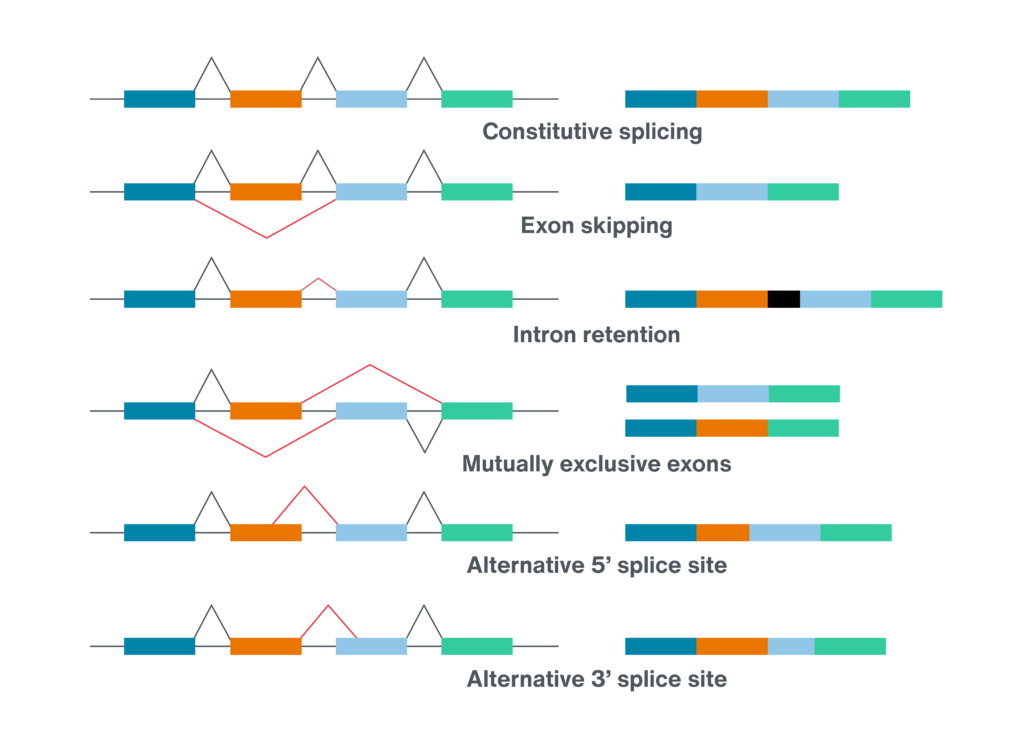
Oxford Nanopore Technologies. Six alternative splicing categories. https://nanoporetech.com/applications/investigations/splice-variation
Rong, S. et al. Large-scale functional screen identifies genetic variants with splicing effects in modern and archaic humans. Proceedings of the National Academy of Sciences 120, (2023). https://www.pnas.org/doi/full/10.1073/pnas.2218308120
Written by Kelley Paris. Contact the author at keparis@davidson.edu
Read more about the author here.
© Copyright 2022 Department of Biology, Davidson College, Davidson, NC 28036
Interesting article, Kelley. Before exposure to Genomics and this paper, I always assumed introns had little significance since they were spliced out and not expressed. However, they are much more important than I originally thought. It’s crazy to think that processes like splicing introns can contribute to much variation in the differentiation of the human species. Looking at the front figure, we, as modern humans, do share many similarities with Neanderthals and Denisovans. I also like that you brought up national selection and how it plays a huge factor in distinguishing these human species due to splicing activities. I’m glad I read this article and improved my knowledge of human evolution because it’s something I knew little about.