The largest Asian genome-wide association study (GWAS) ever conducted helps us better understand the genetic architecture of systemic sclerosis within and across different populations, highlighting the importance of representation in genomic studies.
This web page was produced as an assignment for an undergraduate course at Davidson College. Link to homepage
Scleroderma, also known as systemic sclerosis (SSc), encompasses a group of autoimmune diseases characterized by connective tissue scarring and internal organ fibrosis. Given its significant morbidity and mortality rates and its poorly understood physiological mechanisms, SSc represents a crucial area of study. While the precise causes of SSc remain elusive, it is widely accepted within the scientific community that both genetic and environmental factors contribute to its development.
Previous research groups, such as Radstake et al in 20101, have found locations in our genes, termed “loci”, related to SSc. However, these studies often suffer from disproportionate representation of Caucasian subjects, minimizing the implications for other ethnic groups (Duncan et Al, 2019)2. A recent study takes an exciting step in a new direction by conducting the largest Asian genome-wide association study to date (Ishikawa et al, 2024)3. Researchers aimed to characterize the genetic underpinnings of SSc in East Asian populations. By comparing their findings with previous studies, they sought to determine whether there were unique genetic architectures specific to East Asian populations.
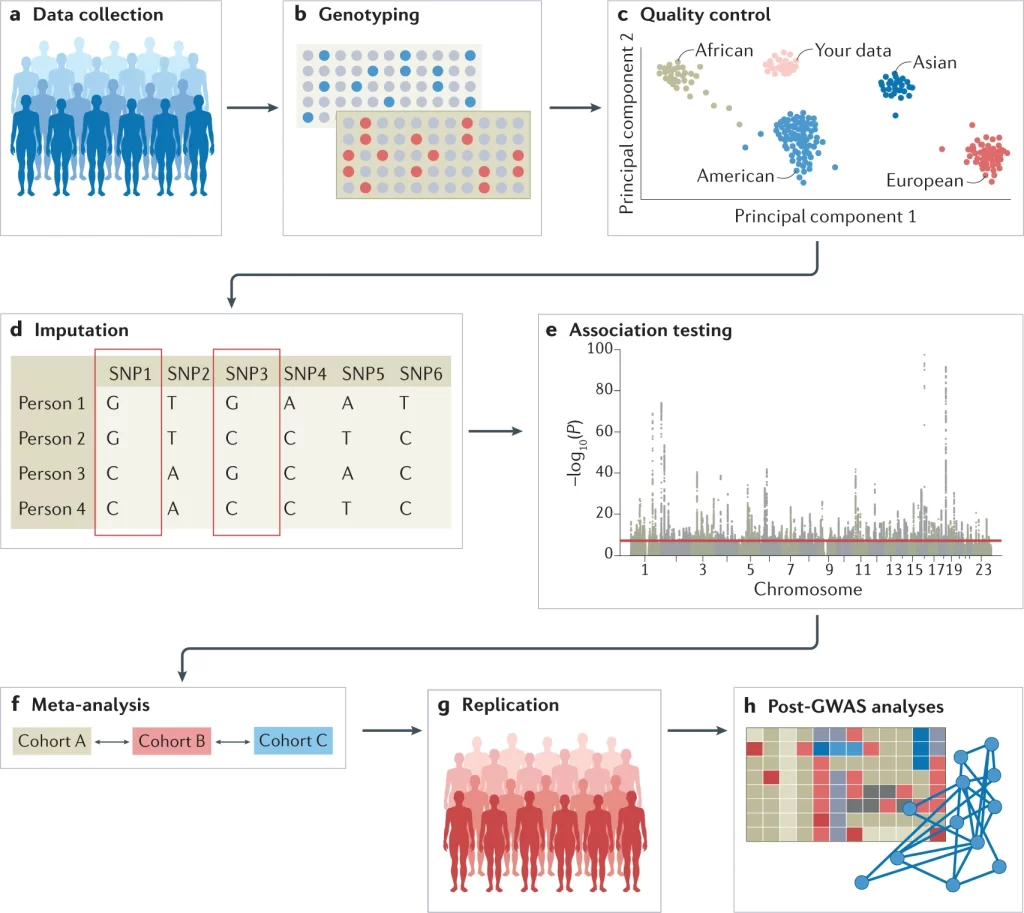
Outline of the major steps, in sequential order, of a GWAS. Credit for image goes to Uffelman et al, 20214.
A genome-wide association study (GWAS) is one of the major analytical techniques used in population genomics. The first step requires collection of the genomes, by some form of DNA sequencing, of two broad categories of individuals: those who have a trait/disease of interest, and those who do not (often called controls). All samples need to be controlled for variables like age and gender to limit the chances of unrelated genetic variety. Then, researchers look for instances of genetic variation in both groups, and compare the frequencies to see if there are locations in the genome where variations happen more frequently in the group who has the specific trait/disease. In theory, this would identify locations of interest in the genome of those who have the disease, which can then be further studied to determine what exactly the relationship is. Genetic variations manifest themselves in many ways, but this study focused on single nucleotide polymorphisms (SNPs). Later on, additional techniques are employed to clean up the data and confirm findings.
Ishikawa et al first sequenced the genomes of 114,026 Japanese individuals. 1,428 of these individuals had SSc, while the remaining 112,599 were controls, or people who did not have SSc. They then used an association analysis – discussed above – to identify a total of five loci related to SSc, three of which were new (RPL31P11-FCRLA, TNFAIP3, AHNAK2). They compared their findings to previous studies and noted that when the same loci had been found across different studies, they had the same effect direction – a term used to describe whether a locus is positively (increases susceptibility) or negatively (decreases susceptibility) linked to a disease. This both validated their findings and suggested that the disease might look similar, in terms of genetic organization, between European and Japanese populations.
Researchers found that variations, specifically SNPs, in the RPL31P11-FCRLA gene had a particularly strong association with SSc in the Japanese population. They studied one particular SNP variant, dubbed rs10917688, and found a link to regulatory gene IRF8, which encodes for the transcription factor of the same name – IRF8. Transcription factors are proteins heavily involved in gene regulation. They are mostly located in the cytoplasm of cells, and upon activation are recruited to the nucleus, where they bind to specific DNA sequences and prevent or/and enhance expression of certain genes. IRF8 is crucial to the development and regulation of immune cells (Moorman et al, 2022)5. Further experiments led researchers to conclude that rs10917688 may impact IRF8’s binding capabilities, specifically in B cells – a type of immune system cell. This finding suggests a possible mechanism for SSc, and also highlights vital loci that may be involved in development of the disease.
Additionally, Ishikawa et al conducted a multi-ethnic GWAS by combining European and Japanese data, focusing on polygenic risk scores. These scores utilize an individual’s genetic variation to assess their predisposition to certain traits or diseases. Previous studies have noted disparities in the predictive power of these scores, particularly in minority groups (Martin et al, 2019)6. However, accuracy improved for Japanese individuals when utilizing scores derived from the combined GWAS dataset.
While this study did not uncover groundbreaking insights into the precise mechanisms of SSc, it made significant contributions to our current understanding. By identifying key players such as IRF8 and B cells, Ishikawa et al laid the groundwork for further investigation. Moreover, their findings underscored the importance of incorporating data from non-European populations, leading to the discovery of novel SSc-related loci and improving current predictive models.
After seeing the value of using a large Asian dataset, the same analysis can and should be conducted with different ethnic groups, such as African-Americans or Latinos. That study might be successful in identifying new loci related to SSc and would only strengthen the predictive power of current statistical measures, even if no discoveries are made. Ishikawa et al, in agreement with previous studies, found that SSc development involves various genes (polygenic), with a large percentage of them being integral to immune function. It would be interesting to further study the interactions between these different genes so that we get closer to a complete understanding of SSc’s molecular pathway. Lastly, the group focused largely on genetic risk factors for SSc, leaving the door wide open for assessing the environmental contributors to the disease.
This article was written by Carlos Jaramillo.
Contact: cajaramillo@davidson.edu
References
- Radstake, Timothy R D J et al. “Genome-wide association study of systemic sclerosis identifies CD247 as a new susceptibility locus.” Nature genetics vol. 42,5 (2010): 426-9. https://doi.org/10.1038/ng.565 ↩︎
- Duncan, L., Shen, H., Gelaye, B. et al. “Analysis of polygenic risk score usage and performance in diverse human populations.” Nat Commun 10, 3328 (2019). https://doi.org/10.1038/s41467-019-11112-0 ↩︎
- Ishikawa, Yuki et al. “GWAS for systemic sclerosis identifies six novel susceptibility loci including one in the Fcγ receptor region.” Nature communications vol. 15,1 319. 31 Jan. 2024, https://doi.org/10.1038/s41467-023-44541-z ↩︎
- Uffelmann, E. et al. “Genome-wide association studies.” Nat Rev Methods Primers 1, 59 (2021). https://doi.org/10.1038/s43586-021-00056-9 ↩︎
- Moorman, H. R., Reategui, Y., Poschel, D. B., & Liu, K. (2022). “IRF8: Mechanism of Action and Health Implications” Cells, 11(17), 2630. https://doi.org/10.3390/cells11172630 ↩︎
- Martin, A. R. et al (2019). Clinical use of current polygenic risk scores may exacerbate health disparities. Nature genetics, 51(4), 584–591. https://doi.org/10.1038/s41588-019-0379-x ↩︎
© Copyright 2022 Department of Biology, Davidson College, Davidson, NC 28036.
Hello, I enjoyed your article! I think you did a great job explaining transcription factors and the significance of key genetic loci such as IRF8. I also appreciate the inclusion of a GWAS description map for the layperson. When reading the article, I scrolled through the chart of the different variants and the effect allele frequency in both Japanese and European populations, and especially noted differences in SLC12A5. I agree with your sentiment in the views section that doing large-scale GWASs for different groups is a great way to find more alleles that could be targeted for treatment. This also increases the likelihood that people of different ancestries will be able to receive therapies that serve to benefit them specifically.
I think this article does a fantastic job of laying the groundwork for this field of Systemic Sclerosis genetics. As a reader, I felt like sufficient background on the disease and what we know about it was given, and it was clear how the newly presented information added to the field. In the views section, I agreed to expand these association studies to include more ethnic groups. I believe it is imperative to include a world-representative population in these studies, as over and over again we have seen that important findings are often missed when a non-representative sample is used. Finally, I appreciated the awareness of the fact that including more ethnic groups may also not reveal new information while highlighting the positive effects of such studies even if this is the case. As a views section, it does a great job of suggesting the remaining questions in the field and providing a rounded perspective of possible findings and implications.