Researchers combined genome-wide association studies in hopes of gaining insight into how much certain gene variations can affect someone’s decision to be the life of the party or stay sober. Their meta-analysis identified the ancestry-specific genetic architecture of alcohol consumption in Japanese cohorts.
Disclaimer: “This web page was produced as an assignment for an undergraduate course at Davidson College.”
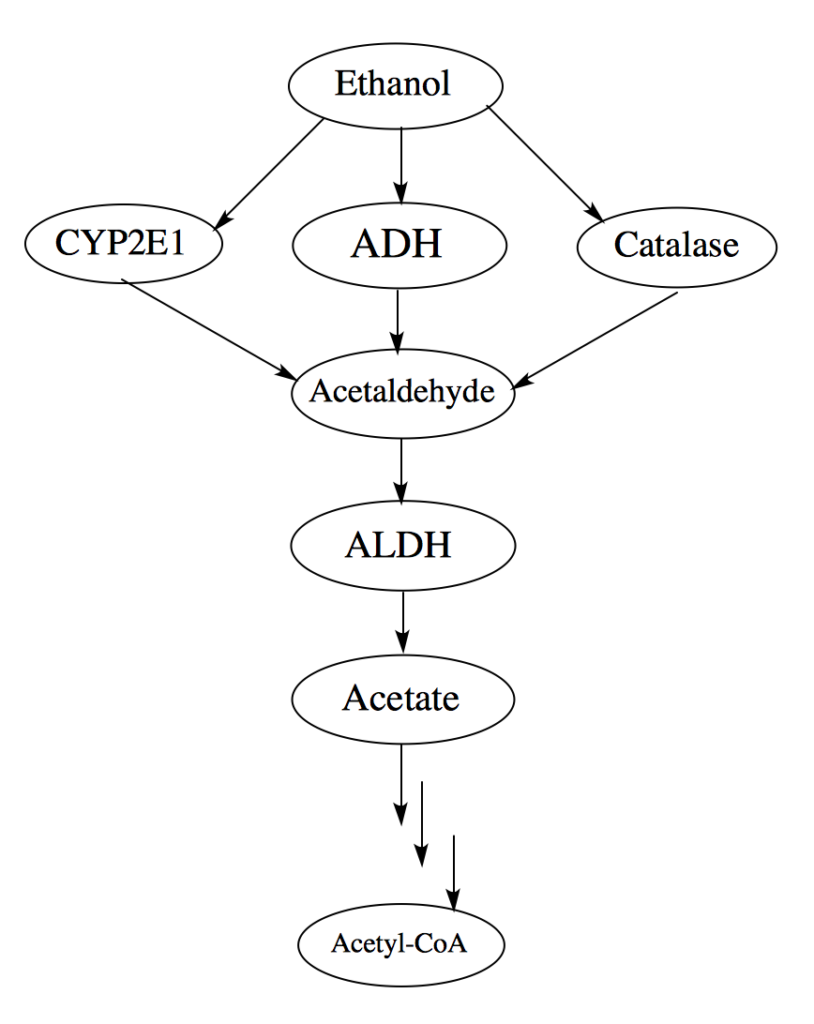
Alcohol consumption can be detrimental to your body, even at moderate levels that don’t interfere with daily life (Rehm 2011). Drinking alcohol, especially in large quantities on a single occasion, can disrupt the way your brain, heart, liver, pancreas, and immune system work. Consuming alcohol puts you at risk for organ damage and certain cancers, especially esophageal cancer (Brooks et al. 2009). Given the harmful effects of alcohol, minimizing your alcohol consumption could reduce your chances of developing these health problems. How each person metabolizes alcohol might also affect the extent to which they experience the harmful effects. The main method through which alcohol is metabolized, known as the oxidative pathway, is diagramed in Figure 1. The compound we call alcohol, ethanol, is metabolized through the oxidative pathway by two liver enzymes, alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH), it’s transformed into different compounds, including acetaldehyde and acetate. If the pathway isn’t working properly, these compounds can accumulate to toxic levels. The enzyme ALDH changes acetaldehyde, an extremely toxic compound, to acetate, a more tolerable compound, through this pathway (Zakhari 2006). Changes to the ALDH enzyme affect how efficiently acetaldehyde is transformed into acetate and the negative effects you’ll experience when drinking alcohol. Slight variations in the parts of the genome that code for the ALDH enzyme are normal, but certain variations can code for a faulty enzyme that can’t convert acetaldehyde to acetate quickly enough or at all, which causes nausea, facial flushing, headache, heart palpitations, and general feelings of sickness.
Figure 1: Flowchart of the pathway for ethanol metabolism. Changes to the ALDH enzyme cause the accumulation of acetaldehyde. Credit to a student blog post from Tufts University.
Previous genome-wide association studies (GWAS) found that a particular variant in the ALDH2 enzyme, labeled “rs671”, codes for an inactive ALDH2 enzyme, thus preventing the conversion of acetaldehyde to acetate (Koyanagi et al. 2024). Individuals with one copy(heterozygous) or two copies (homozygous) of the rs671 variant experience a rapid accumulation of acetaldehyde in their blood after consuming alcohol and the uncomfortable symptoms associated with acetaldehyde toxicity. This variant has been found more frequently in individuals of East Asian ancestry and its symptoms have coined the term “Asian Flush”(Brooks et al. 2009). Koyanagi et al. combined the available studies on the rs671 variant to draw a possible conclusion about the association between alcohol consumption and the rs671 genotype.
Woman of East Asian ancestry displaying the symptoms of acetaldehyde toxicity, “Asian Flush,” after consuming alcohol. Image credit to Stephanie Goh from the TallyPress.com
This group of researchers compiled data from six GWAS with 175,672 participants from Japanese cohorts. The study participants were divided into “never drinkers,” including those who almost never drank, and “ever drinkers,” including former and current drinkers. Individuals were further divided by their genotype for the rs671 variant, GG for wild-type, GA for heterozygous, and AA for homozygous in this variant. By combining the findings of the 6 GWAS and analyzing them together, a statistical tool known as meta-analysis, the researchers were able to find associations between significant loci, specific regions of the genome, and the rs671 variant on alcohol consumption. The loci with genome-wide significance contained single nucleotide polymorphisms (SNP), or changes of a single nucleotide base, that may have some influence and interaction with the rs671 variant associated with the phenotype of interest. The data from each study was not sufficient to make conclusions about the influence of the loci determined to be significant, which is why combining their data was so beneficial.
The meta-analysis found seven loci in significant association with drinking status both individually and through interaction with the rs671 variant. GWAS only provides information about the association between a genetic feature and the phenotype of interest, but answers as to whether the associations are beneficial or harmful require further studies. With the results from the meta-analysis, Koyanagi et al. were able to extrapolate their findings and make predictions about the effect of this variant on enzymatic function, how long ago it arose in the population, and whether it’s being selected for or against. Making any more conclusive claims would require more thorough whole-genome sequencing–based analysis and/or experimental studies.
The meta-analysis results were also used to speculate on the possible protective effect of homozygous genotype on alcohol consumption and alcohol-related diseases and cancers, which could lead to studies on the hereditability of drinking status and even substance use disorders. Researching this East Asian ancestry-specific variant in alcohol metabolism provided useful information about how loci interactions may alter gene expression and/or protein function to different extents. Only a small percentage of participants in GWAS come from non-European ancestry so highlighting and building on existing Japanese studies is important when it comes to making the field of genomics more equitable. The work of Koyanagi et al. is particularly valuable in the movement toward diversifying the participant pool of genomic studies with populations outside of those of European ancestry.
References:
News and Views by: Andrea Morales Correa, anmoralescorrea@davidson.edu
Primary Article: Genetic architecture of alcohol consumption identified by a genotype-stratified GWAS and impact on esophageal cancer risk in Japanese people
© Copyright 2022 Department of Biology, Davidson College, Davidson, NC 28036.”

Wow, this is really interesting! It’s amazing how much our genes can influence how we react to alcohol. Learning about this gene variant rs671 that’s more common in East Asians and how it affects alcohol metabolism is pretty cool. Although I don’t drink much myself, I do notice that I get super flushed in the face at times. I wonder if there’s any connection to the same gene, even without alcohol involved. I completely agree with the importance of diversifying genomic studies to include a broader range of populations in order to ensure that research findings are applicable and beneficial to everyone. I’m excited to learn more about other genetic factors influencing alcohol reactions and if there are potential treatments or interventions for managing symptoms of the “Asian flush” reaction. Thank you for sharing!
Having previously seen “memes” directed towards people experiencing the “Asian flush”, I never attributed this condition to a gene unable to convert a chemical. You mention the “rs671” variant being found more in East Asian individuals, and I wonder why this is the case. Could this be due purely to evolutionary patterns, or can the environment /culture have a part to play? This variant is possibly highly hereditary, and I wonder the extent to which natural selection will select against this variant being present in such a population in the future? I looked online to find if there were medicines to treat this condition, and most recommendations involved drinking less/not drinking at all. It’s also interesting to know that having just one copy of this “rs671” variant (being heterozygous) is enough to display signs of the “Asian flush”.