“This web page was produced as an assignment for an undergraduate course at Davidson College.” About the author: John Ready
Revolutionary metagenomic analysis of environmental samples reveals 16,530 unknown bacterial and archaeal species that could hold powerful insights into the future of drug synthesis and gene editing.
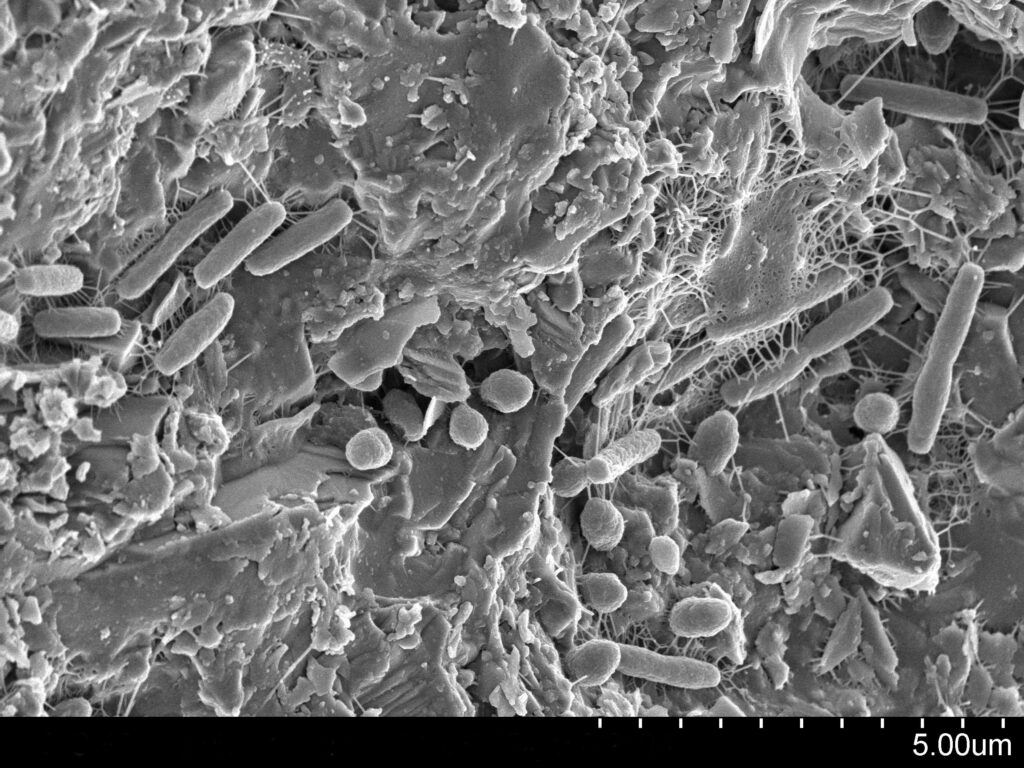
Soil microbiome. Image courtesy of the Lewis Lab at Northeastern University. Image created by Anthony D’Onofrio, William H. Fowle, Eric J. Stewart and Kim Lewis.
Revolutionary metagenomic analysis of environmental samples reveals 16,530 unknown bacterial and archaeal species that could hold powerful insights into the future of drug synthesis and gene editing.
What if I told you that just beneath your feet lies one of the planet’s richest and most diverse ecosystems? You may be skeptical at first, but in a single gram of surface soil, there can exist billions of bacteria and archaea and trillions of viruses1. These bacteria and archaea are known as microbes, or microscopic, single-celled organisms. Don’t let their size fool you, though. These microbes are mighty and play an essential role in soils, maintaining nutrient cycles, soil fertility, and the health of plants2.
Beyond their role in soils, the importance of microbes extends to medicinal purposes as the majority of antibiotic medicine was derived from molecules produced by soil bacteria from the 1940s to 1970s – the golden age of antibiotics. In addition to this advance in medicine, soil bacteria and archaea have contributed to the development of gene-editing technologies such as the CRISPR-Cas9 system. Simply put, this editing tool targets a specific sequence of DNA, that can be cut out or altered to change the way that DNA sequence functions. Despite the significance of soil microbes, an overwhelming majority remained unidentified, owing to the difficulty of cultivating them in a laboratory.
Bin Ma and colleagues noticed this untapped potential and sought to investigate these unknown microbes, or soil microbial dark matter, through genomic methods. Genomics is the study of the entirety of an organism’s genes and it has been utilized to study bacteria since 1995 when the first bacterial genome was sequenced3. Fast forward 29 years later, and there are roughly 500,000 reference genomes for bacterial and archaeal species. Yet there remains a significant portion of unidentified microbial dark matter, so Bin Ma and colleagues began digging it up.
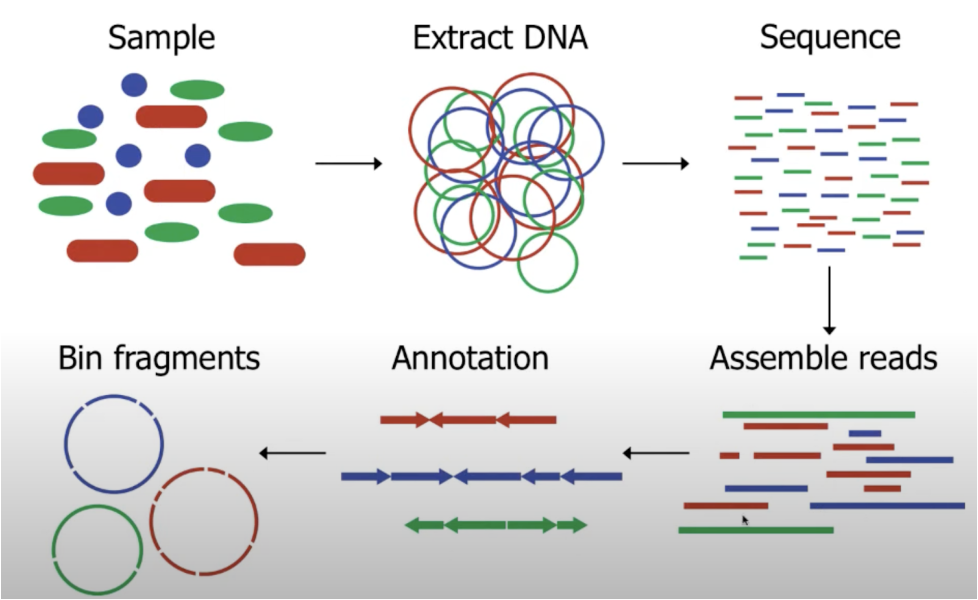
To uncover unknown microbial species the researchers started by creating a genomic catalog, called the SMAG catalog, from 2,941 publicly available soil metagenomes and 363 inhouse soil metagenomes. To create a metagenome, first, an environmental soil sample needs to be collected3. Then from the environmental sample, they extracted and sequenced the DNA, resulting in a metagenome3. The metagenome is essentially all the fragments of DNA from the various bacterial species within that soil sample. These fragments are reads that are sequenced individually from the soil sample.
Metagenome Assembly. Image Courtesy of Canadian Bioinformatics Workshop
From these fragments, Bin Ma and colleagues assembled individual genomes. The assembly process began with assembling initial short read fragments that correspond together into longer continual sequences3. They then utilized a bin sorting technique that takes patterns found in the longer sequences, such as overlaps or matches, and sorts them together. Every bin corresponds to the genome of a species. Eventually, each bin fills up with a sufficient number of sequences to piece together whole genomes. The researchers were left with 40,039 metagenome-assembled-genomes (MAGs), greatly surpassing previous metagenomic soil microbe studies.
From these 40,039 reconstructed microbial genomes, the researchers made some astonishing discoveries. Most notably they identified 16,530 unknown species-level genome bins (uSGBs). The unknown species took up 78.4% of the total assembled species genomes which include previously identified species helping to expand “the bacterial and archaeal diversity across the tree of life”1. Researchers then delved into the functional aspects of the genome. They began by examining the genome for the biosynthetic gene clusters (BGCs), proteins important for metabolic processes, that could be useful for the fields of biotechnology and medicine. Historically, natural products from soil microbes such as BGCs have been a mine for antibiotics4. This mine has slowed down since the golden age of antibiotics, however, Bin Ma et al. found over 70,081 potential BGCs. The abundance of unearthed BGCs could potentially be the source of new therapeutics and drugs.
Relevant to genomics and the future of genetic editing was the expansion of Cas protein and gene resources with the unknown species’ genomes. Bin Ma et al. “detected 8545 natural CRISPR-Cas genes”1. These proteins and genes aid in the CRISPR CAS9 gene editing system that is used widely in genetics research. The new insights from Bin Ma et al. could help improve this tool in the future.
All in all, this study highlights the creation of a new database that extends what we know about the microbial biodiversity of soil ecosystems. Bin Ma et al. provided 16,530 new genomes of unidentified species that help to diversify the microbial tree of life. Perhaps, more significant to some are the medicinal and genetic editing implications of the study. Given what we know about the importance of soil microbes to the discovery of therapeutic drugs, metagenomic analyses could be the future of drug discovery, helping to combat the increasingly antibiotic-resistant bacterial pathogens. Putting this issue into perspective, a study in China determined that antibiotic-resistant infections contributed to the loss of 145,000 lives in 2019 alone5. Statistics such as this show the far-reaching impact genomics can have in the future. Moreover, the addition of genes and proteins related to the CRISPR Cas9 gene editing system is a crucial genetic resource. These new genes and proteins could help to improve the system, thereby making the repair of harmful mutations and modification of existing genes more feasible. Reflecting on this research, metagenomic analyses of soil microbes driven by genomics could usher in a new era in drug discovery and genetic editing, addressing challenges like antibiotic resistance and enabling innovative medical interventions.
John Ready is a class of 2026 Biology major at Davidson College (email: joready@davidson.edu)
For more info check out John’s about me page
References:
- Ma, B. et al. A genomic catalogue of soil microbiomes boosts mining of biodiversity and genetic resources. Nat Commun 14, 7318 (2023).
- Fierer, N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol 15, 579–590 (2017).
- Setubal, J. C. Metagenome-assembled genomes: concepts, analogies, and challenges. Biophys Rev 13, 905–909 (2021).
- Santana-Pereira, A. L. R. et al. Discovery of Novel Biosynthetic Gene Cluster Diversity From a Soil Metagenomic Library. Frontiers in Microbiology 11, (2020).1.
- Zhang, C., Fu, X., Liu, Y., Zhao, H. & Wang, G. Burden of infectious diseases and bacterial antimicrobial resistance in China: a systematic analysis for the global burden of disease study 2019. The Lancet Regional Health – Western Pacific43, 100972 (2024)
© Copyright 2022 Department of Biology, Davidson College, Davidson, NC 28036.
This was an excellent summary, John! I especially like that you described soil as the “planet’s richest and most diverse ecosystem” because it is so true! The more I learn about the intricacies of the world beneath our feet, the more fascinated I feel, and this paper is no exception. This research is also extremely relevant in the age of genome editing. Without the kind of genomic work seen in this paper, we would not be able to study bacterial immune systems or even discover the CRISPR-Cas systems we have today. We can learn so much from other organisms, and bacteria found in soil are no exception!
Ps. One of my favorite podcast episodes of all time is “The Dirty Drug and the Ice Cream Tub,” an episode of the podcast Radiolab that details the discovery of a novel and incredible drug beginning with a molecule found in a soil sample in Easter Island. If you are interested in all the cool things soil can tell us, this episode is for you!
Awesome work John. I think the title was a good way to draw interest in this material. Using “Dark matter” is what caught my attention. I liked your summary and thought you had a nice and concise summary. I really liked that you included an image of the Bin fragment assembly, it made it easier to understand what exactly was being done to assemble the genomes used in the study.
This study appeared to be highly focused on the future uses for possible discoveries being made now. I think this is pretty typical for genomics papers and liked how you highlighted the benefits of the study in an almost sequential order. It seemed as though you placed the benefits right after the discovery, or data, which gave insight into these new innovations.